[ad_1]
In the early universe, the inflationary model holds that the volume of space expanded exponentially and then slowed down. AI-enhanced HPC (AHPC for short) is starting to expand, creating new spaces in the scientific world that are inaccessible (computationally intractable) using traditional HPC numerical methods.
In the world of numerical computing, drawing lines based on the past is one way to predict the future. Although not always perfect, we often extend the lines to predict how fast supercomputers will run HPC benchmarks in the future. These lines reflect computational efficiency and bottlenecks, which ultimately form future short-term exceptions. The same is true for many other applications: benchmark your code, draw lines, and set reasonable expectations.
The linear universe of HPC is about to enter an inflationary period. The capabilities and scope of HPC will accelerate through the use of generative AI (i.e. LLMs). Hallucinations aside, a well-trained LLM can find relationships and features that are unfamiliar to scientists and engineers. LLMs can recognize “features” in data. Consider a feature such as “speed” that is common to many different types of objects: cars, dogs, computers, and molasses. Each of these has some sort of “speed” associated with them. LLMs recognize “speed” and can make associations, relationships, or analogies between disparate pieces of data. (Example: “Cars are faster than dogs” or “This computer is as slow as molasses.”)
There are “dark features” in data that we don’t know about, and with the right training, LLMs are adept at recognizing and leveraging “dark features” in data – relationships or “features” that are invisible to scientists and engineers, but that are still there.
AI-enhanced HPC uses these dark features to expand the HPC computational space. Often called “surrogate models,” these new tools offer scientists and engineers a shortcut to potential solutions by suggesting optimal candidates. For example, instead of 10,000 paths to a solution, LLMs can narrow the range of feasible solutions by orders of magnitude, making what was once computationally insolvable a solvable problem.
Furthermore, using the underlying model feels like an NP-hard problem: creating the model is computationally expensive, but testing results is often trivial (or at least possible in a much shorter time). We are entering an era of AI-augmented HPC, where AI is used to assist traditional HPC computational domains by providing solutions with less computation or by recommending more tractable optimized solution spaces.
These amazing breakthroughs are happening now. Rather than trying to create large-scale general-purpose AI models like ChatGPT or Llama, AI-augmented HPC seems to be focusing on specialized foundational models designed to address specific scientific domains. Here are three examples of such models:
The limits and impact of AI-enhanced HPC are unknown because scientists and engineers cannot see the “dark featurelessness” that foundational models recognize. Progress will not be linear. As we explain below, early foundational models portend a significant expansion of the field of computational science.
Programmable Biology: EvolutionaryScale ESM3
The holy grail of biological science is the ability to understand and manipulate sequence (DNA), structure (proteins), and function (cells, organs). Each of these areas is an active area of research in its own right. Combining these processes opens up a new era of programmable biology. As with any new technology, there are risks, but the rewards will be new medicines, therapies, and drugs that were previously unattainable.
A new company, EvolutionaryScale, has developed a foundational model for life sciences, ESM3 (EvolutionaryScale Model 3), that has the potential to design biology from first principles, just like machines, microchips, and computer programs. The model is trained on approximately 2.8 billion protein sequences sampled from organisms and biomes, and is a significant update from previous versions.
Bioengineering is a difficult endeavor. Based on the human genome (and others), protein folding attempts to figure out the shape of proteins in their biological environment. The process is computationally intensive, and one of the most successful efforts, AlphaFold, used deep learning to speed up the process.
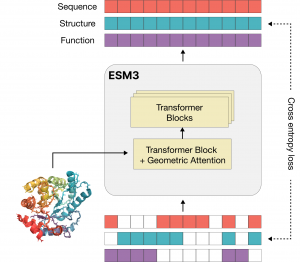
EvolutionaryScale has released a new preprint (currently in preview, awaiting submission to bioRxiv) that describes the generation of a novel green fluorescent protein (GFP) as a proof of concept. Fluorescent proteins are responsible for the glowing colors of jellyfish and corals and are important tools in modern biotechnology. The new protein identified in ESM3 shares only 58% sequence similarity with the closest known naturally fluorescent protein, yet fluoresces as brightly as native GFP. The process is described in detail in the company’s blog:
Generating a new GFP by pure chance (or trial and error) from the vast number of sequences and structures available would be virtually impossible. EvolutionaryScale states that “the rate of diversification of GFPs found in nature suggests that the generation of this new fluorescent protein would be equivalent to simulating more than 500 million years of evolution.”
In their introductory blog, EvolutionaryScale mentions safety and responsible development: Indeed, just as foundational models like E3M3 are called upon to create new candidates for cancer treatment, they may also be called upon to create more lethal agents than currently known. As foundational models are improved and disseminated, AI safety will become increasingly important.
EvolutionaryScale is committed to open development, making its weights and code available on GitHub, and it also lists eight independent research efforts that use its open ESM model.
Weather and climate prediction: Microsoft ClimaX
Another example of AI-enhanced HPC is the Microsoft ClimaX model. Available as open source, the ClimaX model is the first foundational model trained for weather and climate science.
State-of-the-art numerical weather and climate models are based on the simulation of large systems of differential equations that relate energy and material flows based on known physics of various Earth-based systems. As is often the case, such a huge amount of calculations requires large HPC systems. Although these numerical models have been successful, their resolution is often limited by state-of-the-art underlying hardware. Machine learning (ML) models can offer an alternative that benefits from the scale of both data and computation. Recent attempts to scale up deep learning systems for short-term and medium-range weather forecasting have been successful. However, most ML models are trained for specific predictive tasks on specific datasets and lack the general-purpose utility required for weather and climate modeling.
Unlike many Text-Based Transformers (LLMs), ClimaX is based on an improved Vision Transformer (ViT) model from Google Research. ViT was originally developed to process image data, but was later adapted to predict the weather.
ClimaX can be fine-tuned for different forecasting tasks to serve different applications and outperforms state-of-the-art forecasting systems in several benchmarks: for example, using the same ERA5 data, even at medium resolution, ClimaX performs as well as or better than the IFS (Integrated Forecast System, Global Numerical Weather Prediction System).
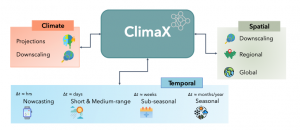
ClimaX is built as a foundational model for all weather and climate modeling tasks. On the meteorological side, these tasks include standard forecasts at different resolutions for different lead-time ranges, both at global and regional scales. On the climatic side, standard tasks include producing long-term forecasts and obtaining downscaling results from lower-resolution model outputs.
Searching for COVID-19 variants at Argonne
Another successful example of domain-specific foundational models was demonstrated by a team of scientists and collaborators at the U.S. Department of Energy (DOE) Argonne National Laboratory, who developed an LLM to aid in the discovery of SARS-CoV-2 variants.
All viruses, like COVID-19, evolve as they multiply (using the host cell’s machinery). With each generation, mutations occur, generating new variants. Many of these variants do not exhibit additional activity, but some can be more deadly and contagious than the original virus. When a particular variant is considered more dangerous or harmful, it is classified as a Variant of Concern (VOC). Predicting these VOCs is difficult because the possible variants are so large. In fact, the key is to predict which variants may be problematic.
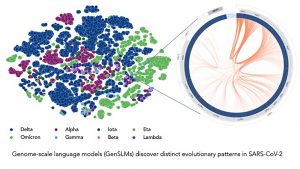
Using Argonne National Laboratory’s supercomputing and AI resources, researchers developed and applied the LLM model to track how the virus mutates into more dangerous or more contagious variants. The Argonne team and collaborators created the first genome-scale language model (GenSLM) that can analyze COVID-19 genes to rapidly identify VOCs. Trained on a year’s worth of SARS-CoV-2 genomic data, the model can infer differences between different viral strains of the virus. Additionally, GenSLM is the first whole-genome-scale foundational model that can be modified and applied to other predictive tasks similar to VOC identification.
Until now, without GenSLM, identifying VOCs would require looking at every protein individually and mapping each mutation to see if it had a mutation of interest. This process is labor-intensive and time-consuming, but GenSLM helps make this process easier.
This figure demonstrates that the GenSLM model is able to infer the distinction between different virus strains.
The research team, led by computational biologist Arvind Ramanathan, included colleagues from Argonne National Laboratory, as well as collaborators from the University of Chicago, NVIDIA, Cerebras, University of Illinois at Chicago, Northern Illinois University, California Institute of Technology, New York University, and Technical University of Munich. The full research can be found in the paper “GenSLMs: Genome-Scale Language Models Reveal Evolutionary Dynamics of SARS-CoV-2.” The project was awarded the 2022 Gordon Bell Special High Performance Computing-Based COVID-19 Research Award for its novel methodology to rapidly identify viral evolution.
Scientific exaggeration
All three of these examples significantly broaden the horizons of their respective domains. Currently, building and running LLM foundational models is still a specialized task. Despite hardware availability, it becomes easier for domain experts to create new and enhanced models. These foundational models recognize the “dark features” of a particular domain and open up science and engineering to new vistas. The world of science and technology is about to get even bigger.
[ad_2]
Source link